Many students feel that, this is one of the most difficult to explain topic in Immunology. Here, we will try to explain the process and clinical relevance of V(D)J or Somatic recombination in simple and interactive way.
Question your mind
Humans only have about 25,000 genes. Since, an antibody made to bind one antigen cannot bind to the other, we need (and we do have) around 100 millions of different antiboies to bind to different antibodies (to protect us from every possible invaders). This antigen specificity of antibody is called idiotype and is determined by the “Variable” terminal of the immunoglobulins. So, if each heavy or light chain protein were encoded by a different gene – how can we produce 100 millions of antibodies from just 25,000 genes.
Answer – Generation of B cell Receptor Diversity
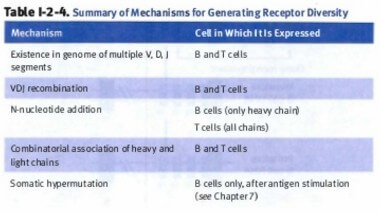
There are 4 mechanisms in which B cells generate receptor diversity:
- Somatic recombination:
- VDJ recombination for heavy chain
- VJ recombination (No “D” segment) for light chain
- Pairing of various light chains with heavy chains
- Junctional diversity (nucleotide addition via Tdt)
- Somatic hypermutation (single point mutation in antibody idiotype)
Before we proceed with discussion of somatic recombination, make sure that you know:
‘Y’
- Immunoglobulin is like an alphabet “Y” within quotation marks. It is made up of 2 identical Heavy chains and 2 identical Light chains.
- The quotation mark represents the light chain and the Y alphabet represent the heavy chain.
- Variable region (V) at the antigen binding region (determines idiotype) is located at head end and the constant region (C) towards the tail end responsible for binding of antibody to cells and complement.
For generation of B cell receptor diversity, we are concerned with the “V” region of antibody here.
Why is somatic recombination called “somatic recombination” ?
This is because it occurs in the somatic cells and not the germline cells.
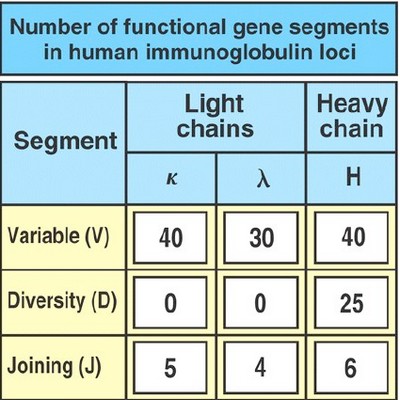
Germ-line DNA on Progenitor B cells provide H and L chain locus – directing the synthesis of Heavy and Light chain respectively. They have V, (D) and J segments in un-rearranged form.
- H chain (chromosome 14) locus has 3 regions – V, D, and J
- L chain (chromosome 2 for κ and 22 for λ) locus has 2 regions – V and J (no “D”)
VDJ rearrangement occurs during the maturation of B cells.
- VDJ rearrangement on ‘H’ chain occurs in Pro-B cells to produce Heavy chain.
- VJ rearrangement on ‘L’ chain occurs in Precursor B cells to produce Light chain.
After the re-arrangement, the B cells are now called Immature B cells.
Trascription of Immature B cell DNA to RNA followed by RNA splicing of introns occur. This brings the Combined V(D)J together with “C” segment that encodes for Constant region of heavy or light chains and forms mRNA.
This is followed by Translation to produce proteins – specific heavy chain and light chain.
V(D)J Rerrangement
Let’s start with the full forms for the “V”, “D” and “J” segments:
- Variable (V) segment
- Diversity (D) segment
- Joining (J) segment
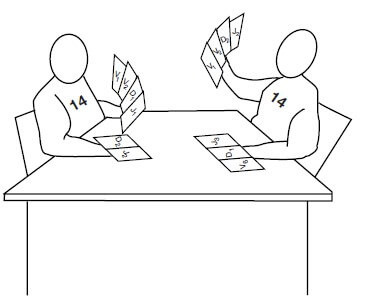
Now, let us simulate a card game between player no. 14 from two different teams (chromosomes).
- Each players have several variety of V, D and J printed (example: V1, V2, D3, J2, etc.) on the cards. Both have the same C printed card.
- Each player tries to rearrange 4 cards each from V, D and J and C (gene segments) until it finds an arrangement that works.
- The first player (chromosome) to do wins and the losing player chromosome stops.
- The players first choose one each of the possible D and J cards and keep them together (by deleting the DNA sequences in between them).
- DJ rearrangement occurs first
- Then one of the many V cards is chosen, and this “card” is kept together with the D and J cards previously matched (again by deleting the DNA in between).
- VDJ rearrangement occurs second
- VDJ-C rearrangement occurs at last
- The player to make these arrangement first, shows the card to the referee and the referee analyzes if the arrangement works or not. If the arrangement is wrong, the referee asks the player to STOP (protein translation stops when ribosome meets one of the three stop codons). Hence, the result of this arrangement is useless.
- In fact, you can calculate that each player only has about 1 chance in 9 of assembling a winning combination of gene segments that will produce a full‐length Hc protein. Immunologists call such a combination of gene segments a productive rearrangement.
- If one of the chromosomes (players) that is playing this game ends up with a productive rearrangement, that chromosome (player) is used to construct the winning Hc protein.
- This heavy chain protein is then transported to the cell surface, where it signals to the losing player (chromosome) that the game is over and the he stops.
- Allelic exclusion: Once a functional product has been achieved by one of the rearrangements, the cell shuts off the rearrangement and expression of the other allele on the homologous chromosome.
This card game simulation also tells that:
- D-J recombination occurs first
- V-DJ recombination occurs second
- VDJ-C recombination occurs last
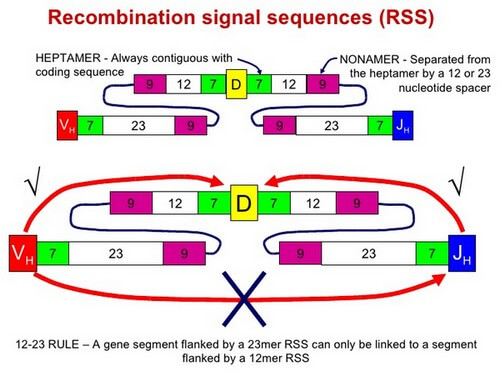
RSS and Rag Genes
Each gene segment (V, D, and J) has an adjacent Recombination Signal Sequence (RSS)
- at the 3′ end of each V segment
- at both ends of each D segment
- at the 3′ end of each J segment
12/23 rule
The configuration prouced by RSS acts as a target for Recombinases. These are recognized by two proteins encoded by two Recombination Activating Genes:
- RAG-1 and
- RAG-2
The RAG-1 and RAG-2 proteins cut through both strands of DNA at the RSS forming double-stranded breaks (DSBs).
The cut ends are stitched together (ligated) to form:
- a coding joint (D-J or V-DJ for heavy chains; V-J for light chains)
- a signal joint (usually a loop of DNA deleting all the intervening DNA initially present between the 2 gene segments chosen)
Other Mechanisms of Generating Diversity
Junctional Diversity
When does it occur?
- During heavy chain gene segment rearrangement process. It doesn’t occur during light chain rearrangement.
Who does it?
- Tdt (terminal dexoyribonucleotidyl transferase) enzyme
How it does?
- It randomly inserts bases at the junctions of V, D and J segments; random in the sense – it can do this without a template on complementary strand.
This generates even more diversity than the random combination of V, D and J segments alone.
Pairing of various heavy and light chains
One can multiply the number of different possible heavy chains by the number of different possible light chains to yield the total number of possible idiotypes that can be generated.
Somatic hypermutation
When does it occur?
- Only when B cells are engaged in response to antigen
What does it do?
- Single point mutation only in the antibody genes – specifically V segment (variable region)
- This leads to increased affinity (antigen binding strength).
- Because binding antigen serves as the first signal for proliferation, over time, clones of cells with higher receptor affinity will begin to predominate the germinal center – this process of clonal selection is known as affinity maturity.
Where does all this happen?
- B cells in the lymph nodes
Isotype switching: This phenomenon is opposite of Somatic hypermutation. During class switch recombination, exchange of one constant region for another occurs, but the heavy chain variable region remains same. This allows effector function of antibody molecule to be changed while maintaining antigen binding.
- Example: switching of IgM to IgE so that it can bind to the mast cells
B cell vs T cell Receptor Diversity Generation
B-cell signal transduction molecules comprises of: Ig-α, Ig-β, CD-19 and CD-21
B-cell receptor (BCR) comprises of 2 heavy and 2 light chains.
In contrast to B cells,
T-cell signal transduction molecule is CD3 and
T-cell receptor (TCR) comprises single α and β chains.
α chain rearrangement is similar to Light chain rearrangement in B cells.
β chain rearrangement is similar to Heavy chain rearrangement in T cells.
Clinical Relevance and Correlation
Tdt
- Tdt is used as a marker for early stage T-cell and B-cell development in Acute Lymphoblastic Leukemia (ALL).
- Myeloid cells stain negatively for Tdt.
- Absent in fetal liver hematopoietic stem cells, which impairs junctional diversity in B cells during the fetal period.
- Tdt+ lymphoid precursors are present in benign pediatric lymph nodes causing possible confusion with ALL.
Omenn Syndrome
- Autosomal recessive disease
- Caused by missense (incomplete) mutation in rag-1 and rag-2 genes – hence, rag enzymes retain partial activity allowing some VDJ combination
- There is releases few lymphocytes, but overall there is lymphopenia
- Peripheral lymphocytes escape to proliferate clonally (homeostatic proliferation) which are regulated by regulatory T cells (T reg) cells to inhibit autoreactive clonal T cell proliferation
- In Omenn syndrome – even T reg cells are also deficient allowing the generation of autoreactive T cells which become hyperreactive and secrete Th2 cytokines like IL-4 and IL-5.
- IL-4 cause isotype switching in the escaped remaining scant B cells to make IgE
- IL-5 tend to eject out eosinophils from the bone marrow leading to eczema, Hyper-IgE syndrome and hypereosinophilia
Omen syndrome – Part of Rag1 and Rag2 gene ommited
- Lymphopenia (immunodeficiency) + Exfoliative erythroderma + Diarrhea + Hepatosplenomegaly
Severe Combined Immunodeficiency (SCID)
- Autosomal recessive disease (however, SCID is most commonly inherited as X-linked disorder)
- Caused by null (complete) mutation in rag-1 or rag-2 genes – hence, there is no rag enzyme activity.
- There is total lack of B and T cells.