Let’s unveil the mathematics behind the relation between:
- Prevalence (Pre-test probability)
- Predictive value (Post-test probability)
- Sensitivity
- Specificity
SYMBOLS:
Total screened population = n
Prevalence = P
Sensitivity = Sn
Specificity = Sp
True positive = TP
True negative = TN
False positive = FP
False negative = FN
With disease = D+
Without disease = D-
Positive predictive value = PPV
Negative predictive value = NPV
Since, we calculate all these in percentage, lets assume n = 1
If n = 1,
Total diseased = D+ = P
Total without disease = D- = 1-P
WE WILL PLAY WITH THESE SYMBOLS:
Sn = TP rate = TP/D+
or, TP = Sn X D+ = SnXP
Sp = TN rate = TN/D-
or, TN = Sp X D – = SpX(1-P)
1-Sn = (D+ – TP)/D+ = FN/D+ = FN/P
or, FN = (1-Sn)XP
1-Sp = (D- – TN)/D- = FP/D- = FP/(1-P)
or, FP = (1-Sp)(1-P)
Mnemonic:
Just remember, prevalence (P) is there in all.
a. TP = + X + (truly positive)
b. TN = + X – (one positive and one negative)
c. FN = – X + (one positive and one negative)
d. FP = – X – (both negative by product is falsely positive)
FN has letter N, Sensitivity has letter N and there will be minus (-) on Sensitivity: FN = (1-Sn)XP — one negative, one positive
FP has letter P, Specificity and Prevalence both have letter P and there will be minus (-) on both: FP = (1-Sp) X (1-P) — both negative
Remember the other two, these are opposite. While FN relates to Sn, TN relates to Sp and while FP relates to Sp, TP relates to Sn.
TN = Sp X (1-P) — one positive one negative
TP = Sn X P — both positive
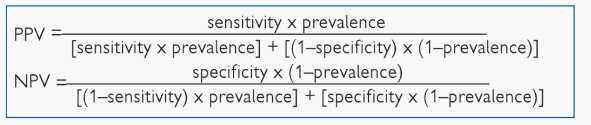
PPV = TP/(TP+FP)
= SnXP/[SnXP + (1-Sp)(1-P)]
NPV = TN/(TN+FN)
= SpX(1-P)/[SpX(1-P) + (1-Sn)XP]
You are often tested on exams with numerical questions. This is how, I try to figure out the Bayism in PPV and NPV when forgotten.
I have three genes as a biomarker for CRC disease and need to combine their diagnostic values?