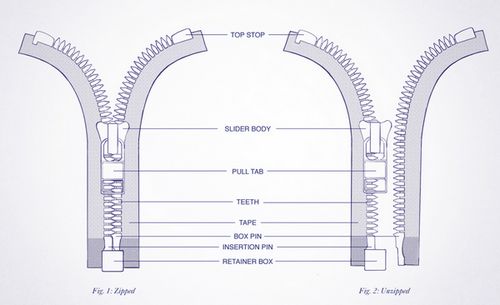
Imagine DNA as a zipper
Prokaryotic DNA as a zipper with single slider (single origin of replication) and Eukaryotic DNA as a zipper with two sliders (multiple origin of replication).
Zipper teeth: Purines and pyrimidine bases
- Complementary teeth pair: Complementary base pairs attached by hydrogen bonds
Top stops: Origin of replications.
Slider handle (when opening zipper): Helicase.
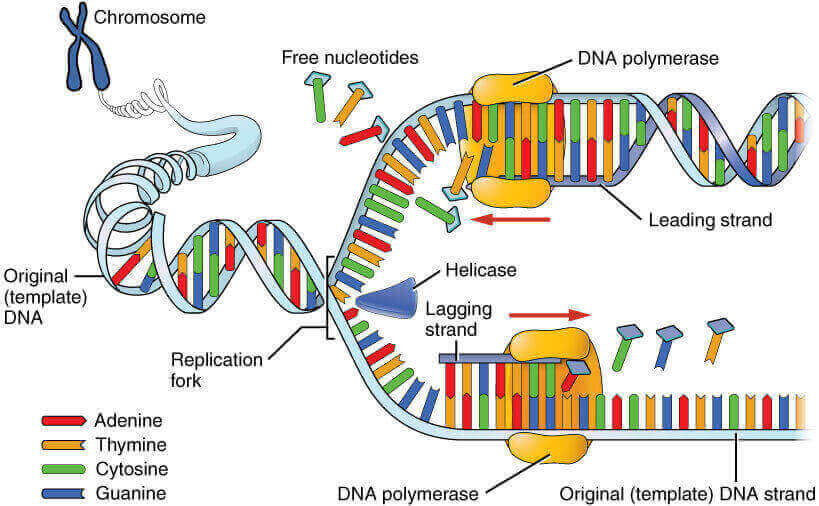
Opening of zipper: Unwinding of DNA and formation of 2 replication forks.
- DNA topoisomerase II (a.k.a DNA gyrase in prokaryotes) removes positive supercoils ahead of advancing replication.
- Topoisomerase I nicks DNA, relieving torsional tension of the replicating helix.
- Topoisomerase II introduces the double strand break to avoid DNA tangle.
Side tapes supporting Zipper teeth: Single-stranded DNA-binding protein (SSB) that supports and prevents reassociation and degradation of single stranded DNA by nucleases.
Retainer box and insertion pin: RNA primer synthesized by Primase
Slider handle (when closing zipper): DNA polymerase III in eukaryotes and DNA polymerase α + δ
Other Eukaryotic DNA polymerases:
DNA polymerase γ: replicates mitochondrial DNA
DNA polymerase β and ε: participate in DNA repair
Closing of zipper: Synthesis of new strand in 5’→3′ direction beginning at 3′ end of RNA primer.
- New zipper closes smoothly: Leading strand
- Broken zipper that gets stuck several times before closing: Lagging strand (Okazaki fragments are synthesized discontinuously and later connected by enzyme ligase).
Zipper handle (DNA polymerase) as exonuclease:
- When closing zipper: 5’→3′ exonuclease (Removes RNA primers) – DNA polymerase I (prokaryotes) and RNAse H (eukaryotes)
- When opening zipper again: 3’→5′ exonuclease (Proofreading new strand) – DNA polymerase
DNA repair:
Mismatch repair (during replication): by MSH2 and MLH1 repair genes.
Thymidine dimers (UV radiation): Excision endonuclease/Excinuclease (deficient in Xeroderma pigmentosum) removes defective oligonucleotide, DNA polymerase fills the gap and DNA ligase seals the nick in repaired strand. This is nucleotide excision repair.
Cytosine deamination (Cytosine converted to Uracil spontaneously or by heat):
- Uracil glycolase removes uracil (base excision).
- AP endonuclease removes empty strand segment.
- DNA polymerase fills the gap
- DNA ligase seals the nick in repaired strand.

He is the section editor of Orthopedics in Epomedicine. He searches for and share simpler ways to make complicated medical topics simple. He also loves writing poetry, listening and playing music.


The Double Helix is wrong: there is no mutual twisting of the two strands — they run in parallel (of course they are antiparallel).
Rosalind Franklin was pretty much right. As Aaron Klug wrote,
Her notebooks for the winter of 1952—53 show her considering a variety of structures including sheets, rods made of two chains running in opposite directions with interdigitated bases and also a pseudo-helical structure with non-equivalent phosphate groups which looked like a figure of eight in projection.